![back to Spectroscopy Ninja [Spectroscopy Ninja]](../Spectroscopy-Ninja_lightblue_230x100.png)
Spekwin32
About | Manual | News | Download | Copyright | Forum | Links | Testimonials | Contact | Imprint
About | Manual | News | Download | Copyright | Forum | Links | Testimonials | Contact | Imprint
Share this page on:
- Main Window
- File
- Spectra General
- Calculations
- Fluorescence/EEM
- Gaussfit
- Plot/Options
- Help
Information
Normalize
Baseline Correction
Advanced Baselining
Spike Removal
Play
Smooth
Advanced Smoothing
Find Peaks & FWHM
Integration
Cut off Spectrum Part
Sort
remove
History
News
Download
License Order
Copyright
Links
User Forum
Testimonials
Contact
Spectra menu
1.
Menu Item [General]
- Here you
can change all parameters important for the display of spectra. Further,
the absorption path length can be set and the sample concentration
can be calculated from molecular weight, sample weight and solvent volume.
- All values
displayed and entries made belong to the current spectrum, selected
in the drop down list above. The current spectrum displays as a thicker
line in the Graph window.
- Legend
Title: can be changed in the upper drop down list for each spectrum.
With <Up> and <Down> keys you can scroll through the list
of Legend Titles, the same works with the dialog's [<= Back] and
[Forward=>] buttons. Of course, spectra can also be directly selected
in the upper drop down list. All entries are automatically updated on
making changes to the selected spectrum. With [Replace by filename]
the legend title of the selected spectrum will be replaced by its filename.
With [Change all titles...] you can work on all legend titles in a single window. The '#' character may be used as a wildcard with search/replace.There, it is also possible to replace the legend texts by their file names.
- Line
Width: can be set for any spectrum in relation to the general line
width which is changed in menu items "Thicker
lines" and "Thinner
Lines". For exporting as graphics and printing, it is recommended
to select line widths from 2 to 5, depending on the outcome of the printed
graph.
- Temperature:
For some spectral file types (*.ggg, *.gdm, *.gkl, *.gpo) the measuring
temperature is shown. You may enter your own values.
Wavelength: For some spectral file types (*.sp, *.ggg, *.prn, *.spe) the wavelength is shown (excitation wavelength for fluorescence spectra, detection wavelength for excitation spectra). You may enter your own values. The only commercial file type having its wavelength inside is *.sp (Perkin Elmer fluorescence spectra).
- Line
Type: there are three line types possible which can be set for any
spectrum separately.
- Line
Colour: the line colour of the current spectrum is shown in the
vertical colour field on the right. There are 14 different colours in
the palette which cycle infinitely. The line colour can be set for each
spectrum separately with the button [Change colour =>]. There you
can also define your own colours.
- Absorption
Path Length: standard value is 1cm. Changes have an effect on y
axis scaling with
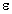 and
and 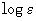 ,
as well as for calculation of concentration via the molar decadic absorption
coefficient with the menu item Concentration.
,
as well as for calculation of concentration via the molar decadic absorption
coefficient with the menu item Concentration.
- Concentration:
standard value is 0 mol/l. For y axis scaling with
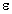 and
and 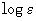 a concentration
a concentration  has
to be entered. After entering values for molecular weight, sample weigth
and solvent volume, the concentration can be calculate with the button
[Calculate].
has
to be entered. After entering values for molecular weight, sample weigth
and solvent volume, the concentration can be calculate with the button
[Calculate].
2.
Menu Item [Information]
- For the spectrum selected in the upper drop down list the following properties are shown: filepath, filename, date, description, comment1, comment2, spectrum type, solvent, absorption path length, concentration, wavelength, temperature, start wavelength, end wavelength, step width, number of data points.
- The text
in the lower part may be copied.
3.
Menu Item [Normalize], (Peak) or (Area)
- Peak:
Normalization in this context means to set the maximum value of the
spectrum to 1,
Area: to set the area of a certain region to a common value for all spectra.
- Peak: All
spectra are normalized to 1 at their maximum wavelength in the currently
displayed x axis range. To select a certain maximum for normalization,
zoom into a x axis range where this maximum is the highest.
Area: Use the left mouse button to select the region for area normalizaiton. All spectra are normalized to a the same integral value for the peak area in the selected region. The x axis boundaires for this region are shown in the lower status bar.. - Normalization
changes the y values irreversibly and can be done as often as you want.
This function is very helpful for direct comparison of spectra.
4.
Menu Item [Baseline Correction]
- For offset-correcting
of straight baselines that deviate from zero. Call the function and
use the mouse zoom function to select the range which should be set
to zero. The mean value of y values in the selected region will be subtracted
from the spectrum as a constant value.
- Will be
done for all spectra at once.
5.
Menu Item [Advanced Baselining]
- For cases,
where a simple offset baseline correction doesn't work, this menu function
gives advanced possibilities. Select the spectrum for adapting the baseline
in the upper drop down list. The treated spectrum will be added to the
list of spectra. An interactive live preview helps to efficiently adapt
the baseline. Three different baseline types exist currently:
- The linear
option creates a straight baseline, that can be rotated and shiftet
up and down.
- The adaptive
baseline will be most useful for Raman and FTIR spectra, where a slowly
curving baseline has to be subtracted from a spectrum with narrower
peaks. The "coarseness" can be varied, and the result can
be shifted up and down.
- A scattering
solution has a baseline continuously rising towards short wavelength
values. The increase follows a power law, with an exponent depending
on particle sizes. The resulting baseline can be compressed or expanded
vertically and shifted up and down.
- The following
settings are available:
- [treat all spectra] subtracts the created baseline from all loaded spectra.
- [remove original] removes the original spectrum/ spectra.
- [keep legend] lets the original legend text unchanged. If unchecked, " (baseline corrected)" will be added to the legend text(s).
- [create
baseline spectrum] will produce a spectrum from the baseline used.
This can be treated like every spectrum (save, etc.)
6. Menu Item [Spike Removal]
- When using
prolonged exposure times with CCD detectors, artefacts from cosmic radiation
are increasing. These are called cosmic spikes.
- This function
detects spikes and interpolates the spectrum in the range of the spike.
All other spectral data points remain totally unchanged. Spectra without
spikes remain also unchanged. Broad spikes (> 5 data points, quite
rare), are not detected to make sure not to change spectral features.
- Select
the spectrum to be treated in the upper drop down list. The treated
spectrum will be added to the list of spectra.
- Options:
- [Remove Original] removes the original spectrum.
- [Treat all Spectra] for treating all spectra, original spectra will be removed.
- [Backward spike Removal] lets the detection algorithm move over the spectrum from right to left. In this way some more spikes might be detected.
- [Append
"spikes removed"] appends "spikes removed" to
the legend titel of the spectrum.
7.
Menu Item [Play]
- The y values
are transformed to tone pitches and played from left to right via the
case speaker. The playing speed is 50 nm/sec, independent of the resolution
of the spectrum.
- You might
even play real music on selection of appropiate y values and wavelengths.
Examples: "Alle meine Entchen" und
"Bourree"
8.
Menu Item [Smooth]
- Smoothing all spectra at once.
- The algorithm
is equivalent to using Savitsky-Golay smoothing of third polynomial
order from the "Advanced Smoothing" menu item, however smoothing
intensity depends on the resolution of spectra ("interval"
parameter will be kept between 6 and 30 points).
- Smoothed
spectra are added to the list of spectra, the legend titles are appended
by "smoothed". Keep this function in mind to be a suspicious
feature, it means modifying and glossing over the original data.
9.
Menu Item [Advanced Smoothing]
- For better
control of the smoothing behavior. Select the spectrum to be smoothed
in the upper drop down list. The treated spectrum will be added to the
list of spectra. Three different smoothing types exist currently:
- [moving average] smoothes by calculating an average value for every data point from a data range around this point with the selected interval size. A rectangular or triangular shape can be choosen. With the triangular shape, more distant data points are down-weighted.
- [Savitsky-Golay] smoothes by calculating a least-squares polynomial interpolation value for every data point. The interval size and the polynomial order can be choosen.
- [Percentile
filter] smoothes by identifying the selected percentile for the
data points contained in the interval size successively for every
data point. A lower or upper envelope spectrum results upon choosing
the 0% or 100% percentile.
- The following
settings are also available:
- [treat all spectra] smoothes all loaded spectra with the selected parameters.
- [remove
original] removes the original spectrum/ spectra.
10.
Menu Item [Find Peaks & FWHM]
- Select
a spectrum in the upper drop down list. In the lower part, a peak
table will be shown.
- If possible,
the corresponding FWHM (full width at half maximum) values will be shown.
- The peak
finding threshold is variable from 1 - 95%.
- For finding
minima, activate the [ ] inverted checkbox.
- The text
in the lower part may be copied.
11.
Menu Item [Integration]
- After calling
this menu item, use the mouse to zoom into the x axis range to be integrated.
Left and right border of the zoom box are the integral boundaries.
For each spectrum a separate message window is shown, containing the
boundaries, the integral value of the spectrum and the average value
within the selected range.
- Hint:
The oscillatory strength will be shown additionally, if the axis
types wavenumbers and absorption coefficient are selected. Of course,
the value of the oscillatory strength makes sense only for selection
of the first electronic transition for integration. The following formula
is used to calculate the oscillatory strength:
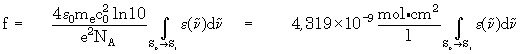
12.
Menu Item [Cut off Spectrum Part]
- For reducing
the spectral range (x scale).
- Select
the spectrum to be cut in the middle drop down list and enter the boundaries.
The treated spectrum will be added to the list of spectra. The new spectral
boundaries will be kept as long as the program runs.
- Options:
- with "all spectra of one type", you can treat all spectra of a certain type simultaneously.
- [remove original(s)] removes the original spectrum/spectra.
- [prepend
"part of:"] prepends "part of:" to the legend
titel of the spectrum/spectra.
13.
Menu Item [Sort]
- For changing
the order of loaded spectra.
- In the
left window, spectra order can be changed manually. Left mouse button
for selection (use SHIFT to select multiple discrete spectra). Insert
selected spectra with the right mouse button (above the spectrum you
are pointing to). With the option [automatically] all spectra can be
sorted automatically by three criteria in both directions:
- <Direction>
can be applied to all spectra (alphabetical and inverse alphabetical
available).
- <Temperature>
only for spectra of our home built spectrometers
- <Wavelength>
for some spectra of our home built spectrometers and Perkin Elmer
*.sp spectra, if measured with a fluorescence spectrometer. For
*.spe files, the laser wavelength will be displayed, if they are
of Raman type.Up and down direction available.
- <Direction>
can be applied to all spectra (alphabetical and inverse alphabetical
available).
14.
Submenu [Remove]
- 13.1.
Menu Item [Remove All]
All loaded spectra are removed. You have to confirm this before removal takes place actually.
- 13.2.
Menu Item [Select for removing]
For removing arbitrary spectra. Select spectra for removing on the left, bring them to the right with the button [>]. With [>>] you can bring all spectra from left to right at once. The same holds with [<] and [<<] for the opposite direction.
With the option "remove all spectra of one type", you can remove all spectra of a certain type at once.
- 13.3.
Menu Item [Remove Last One]
The last spectrum from the list will be removed.